End-to-End Recommendation with MolRec
Molecule Recommender workflow

Exploring the vast and growing landscape of chemical compounds is a significant challenge for researchers, therefore making the development of an effective chemical recommender system not just interesting but crucial. Building such a system has the potential to revolutionize how scientists discover and interact with chemical data, enhancing their research productivity and sparking innovation across chemistry and allied sciences. Previous works on recommendation of chemical compounds has been limited and not widely explored, indicating a significant opportunity for developing systems that can effectively navigate, predict and recommend novel and existing chemical compounds. The gap in the literature underscores the necessity for methods that integrate diverse algorithmic approaches, aiming to improve the discovery and identification process within extensive chemical datasets.
Towards this goal, we introduce a hybrid chemical recommender system which leverages a combination of algorithms, including collaborative filtering, content-based approaches, and Graph Neural Network variational autoencoders to effectively identify and recommend compounds of interest. It focuses on introducing scientific researchers to potentially unknown chemical compounds within large-scale chemical datasets, enhancing discovery and research efficiency in chemistry and related fields.
Our end-to-end recommendation model is outlined in the figure above. In the first stage, we use classical IR methods such as collaborative filtering and content-based approaches to recommend multiple reference molecules to each researcher based on their previous research interests. This is done using a dataset of our own construction which lists molecules researchers have interacted with in their previous papers. The methods in the first stage and MolRec data are described in greater detail at Stage I: Classical Recommendation. In the second stage, we use these reference molecules and a variational autoencoder GNN to recommend novel molecules to the researcher beyond the molecules in the dataset, described in greater detail at Stage II: Deep Generative Model.
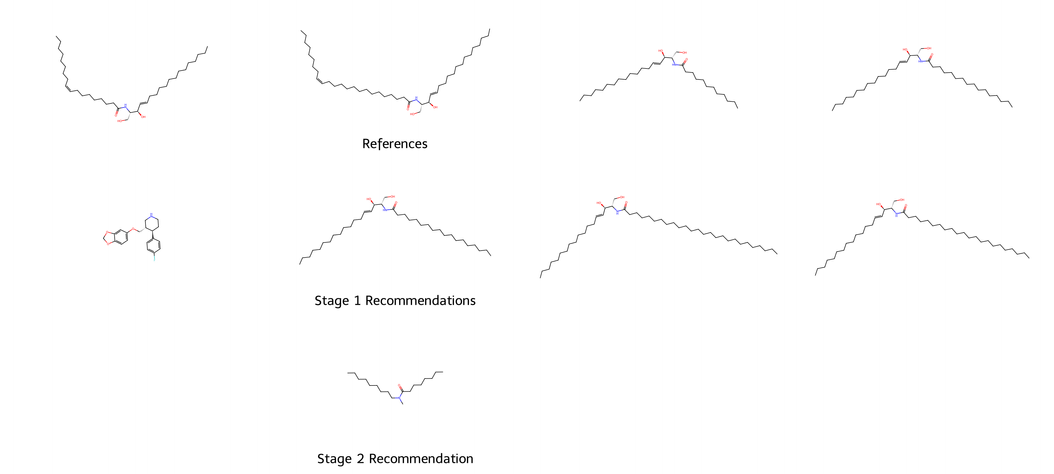
We next show several end-to-end recommendations for selected researchers. Please visit the end-to-end recommendation visualization section of our GitHub or the gallery below for more. In these visualizations, the top row shows reference molecules that the researcher has previously interacted with in their work. The Stage I Classical Recommendation methods use these reference molecules to recommend several molecules from the MolRec data, shown in the middle row. Finally, given the Stage I recommendations, the Stage II VAE generates a novel molecule, which is shown in the third and final row.

Explore more examples
Unlocking the Mysteries of Chemical Compounds: Why It Matters
🌟 The Quest for Discovery: Imagine a world where uncovering new compounds isn't just a laborious task, but an exhilarating journey of exploration. That's the promise held by the development of an effective chemical recommender system. It's not just interesting—it's a game-changer.
💡 Sparking Innovation: Picture this: A tool that revolutionizes how scientists navigate the sea of chemical data. With such a system in place, researchers can soar to new heights of productivity, sparking innovation across chemistry and allied sciences. It's not just about making their jobs easier—it's about unlocking the door to groundbreaking discoveries.
Collaborative Filtering
Given the dataset with columns "user", "compoundID", "rating", and chemical pictures, we use collaborative Filtering to recommend items to users based on their past interactions and similarities with other users. This technique analyzes patterns of user behavior to predict how a user might rate or interact with items they have not yet seen. By leveraging the ratings and compound IDs in the dataset, collaborative filtering identifies users with similar tastes and preferences and recommends items that these similar users have rated highly.
Chemical Semantic Similarities
Semantic Similarity (Content Based) component leverages Chemical semantic similarity based on the ChEBI ontology (ONTO). This involves using DiShIn for calculating distances between entities in the semantic base and employs the Resnik similarity metric to quantify the semantic relatedness between items.
GNN - Chemical Molecular Graphs
We train a generative GNN based on the JTVAE architecture for generation of novel and chemically valid molecules.